Bacterial artificial chromosome BAC: An artificially created bacterial plasmid, which includes all the areas needed for replication (ORI and the needed coding DNA). It copies itself only once or twice inside the host cell, but compared to a yeast artificial chromosome BAC is more stable, easy to transform, easier to clean and grows well in E. coli.
Cap-structure: mRNA is capped. The cap-structure is added to the 5'-end of the pre-mRNA during transcription, and seems to be involved in several aspects of pre-mRNA and mRNA metabolism, in most cases related to identifying the 5'-end of the RNA.
Cloning vector: A sequence to which foreign genetic material has been attached, and which then is transferred into a host cell. Most commonly the vector is a bacterial plasmid or a virus. Forget mathematical vectors or vector graphics, this vector is more of a "vessel". Plasmids can carry inserts up to 10 bp, bacterial artificial chromosomes up to 300 and yeast artificial chromosomes up to 200-2000 bp.
Codon/anticodon: A codon is a base triplet in the mRNA, and anticodon is its counterpart in tRNA. By matching an anticodon to a codon the correct amino acids can be added to the protein chain. Only the two first bases are important, causing a wobble-effect (the third codon can be different, and still the amino acid is recognized correctly). Each protein synthesis starts with a start codon (AUG) and ends in a stop codon (UAG, UGA, UAA).
Cosmid vector: A plasmid to which a cos-area from a lambda-phage (see bacteriophage) has been inserted. The cos area enables packing the cosmid inside a protein coating of the phage. Used commonly when building cDNA-libraries.
DNA ligase: An enzyme which attaches two single strands of DNA together. It does not create a dual-stranded DNA, but only elongates an existing single strand by adding another piece to it. It is used in DNA transcription to close the gaps between Okazaki fragments, and when attaching DNA strands cut with restriction enzymes back together or to a vector. Needs ATP and Mg2+ to work.
DNA polymerase: A DNA polymerase is a cellular or viral polymerase enzyme that synthesizes DNA molecules from their nucleotide building blocks. DNA polymerases are essential for DNA replication, and usually function in pairs while copying one double-stranded DNA molecule into two. DNA polymerases also play key roles in other processes within cells, including DNA repair, genetic recombination, reverse transcription. (Wikipedia)
Expression vector: A vector built specifically for producing proteins. An expression vector includes the needed DNA-insert for coding the protein, a promoter to start the DNA transcription and start and end sites for the actual protein synthesis. Expression vectors are host cell specific.
Exon: A part of a gene which codes proteins. When a finished DNA-strand is spliced, exons are included in the end product. Some exons may be left out to achieve different variations of the gene.
Intron: A part of a gene which does not code any protein, or include functional DNA. Introns are left out when splicing.
 |
| Structure of a nucleotide. (c) Wikipedia |
PCR (polymerase chain reaction): A method to replicate DNA automatically in a test tube using DNA polymerase. It has three stages: DNA denaturation, binding of the primers and DNA synthesis. A simple animation of it can be found from the website of Wiley. Many polymerases are sensitive to the Mg2+ concentration, so it has to be optimized for the polymerase used in the PCR.
Peptide bond: A bond formed between two amino acids in protein synthesis, when a carboxyl group and an amino group bind together and one molecule of water (H20) is lost.
Plasmid: A small, circular dual-strand of bacterial DNA, which is often used as a cloning vector. They replicate independently inside the bacterial cell. Plasmids always have a point of origin (ORI), which is the starting point for the replication. Plasmids altered for use in biotechnics have a cloning site, which includes restriction sites for several different restriction enzymes. Recombinant plasmids also have a selection gene, which is used to distinguish recombinants from wild types.
Polysome: A site for protein synthesis. Polyribosomes (or polysomes) are a cluster of ribosomes, bound to a mRNA molecule. Many ribosomes read one mRNA simultaneously, progressing along the mRNA to synthesize the same protein. (Wikipedia)
Primer: An RNA/DNA sequence serving as a start point for DNA replication. It's 18-22 bp long, and can be designed and built artificially. The polymerase starts replication at the 3'-end of the primer, and copies the opposite strand. Primers are used in PCR to give the polymerase a place to bind to. Primers are "independent" strands, which bind to the DNA to be used as a model, while promoters are sequences in the actual DNA.
Promoter: An organism-specific site where the polymerase enzyme binds and where the DNA- or RNA-replication starts. There are three types of promoters: 1. constitutive promoters, which work all the time in every tissue type in the organism, 2. tissue- or time-specific promoters which work only in some tissue types or during a certain time, 3. inducible promoters, which work only when a certain stimuli is present. (PatentLens) Eucaryotic cells have a promoter called TATA box, located 25 bases upstream from the start point of the transcription. Promoters can control the efficinecy of RNA synthesis.
RNA types: rRNA: ribosomal RNA. Exist in ribosomes, where they hold the mRNA steady and assist in protein synthesis. mRNA: messenger RNA is a copy of genetic information (DNA), which is used as a template for protein synthesis. tRNA: transfer RNA partakes in protein synthesis by matching a base triplet (a codon) to a correct amino acid, which it brings to the synthesis site.
Reading frame: Reading frame means how the DNA/RNA is "split" into three bases for reading. A sequence of AACTGTAC could be read for example as AAC|TGT|AC or as A|ACT|GTA|C. If there are over 50 amino acids between the start and stop codons, the protein has an open reading frame.
 |
| Enzymes used in recombinant DNA technology. (c) Lecture material, origin unknown. |
Replication: A process where a dual-strand DNA is opened, and both strands are used as a template to create a new dual-strand DNA. The process is semiconservative, meaning that both new strands have 50 % of the original genetic material.
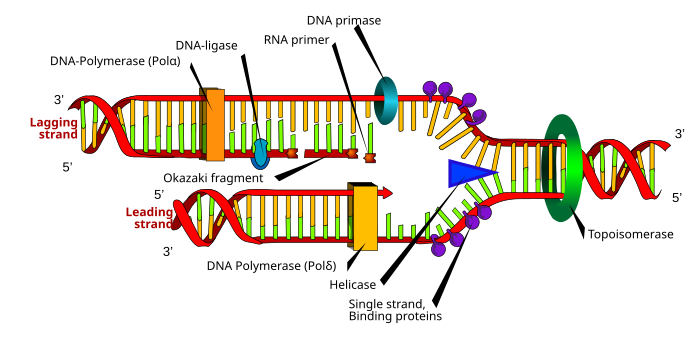 |
| A replication fork. (c) Wikipedia |
Restriction enzyme: An enzyme, which recognizes a specific sequence of DNA and cuts it. The recognized sequence is called a restriction site. Chromosomes and bacterial plasmids have hundreds or thousands of restriction sites for different enzymes. Restriction enzymes are very useful in gene technology, where the genes can be cut from precise points for sequencing or ligating to another chromosome. Restriction endonucleases cut the strand from the middle, while exonucleases cut it from the ends of the strand.
Restriction site: A palindromic site where a restriction enzyme makes the cut. Restriction sites 4 bases long appear approximately once every 300 nucleotides, so enzymes using these cut the DNA into very short strands. Restriction site of 6 bp appear once in 4000 nucleotides, and those with 8 bp very rarely.
RNA structure: One stranded, consists of adenine, uracil, cytocine and guanine. The 3' carbon of the sugar ring has a hydroxyl group (-OH). It may bind on its own, creating loops and hairpins.
Transformation of bacteria: The process of a bacteria accepting pure DNA into its cell. If the inserted DNA includes an ORI site, the DNA will be replicated inside the cell. The plasmid determines how many copies are made. Before transformation the bacteria has to be made competent, i.e. ready to accept DNA. This can be done by CaCl-treatment, which causes the cells to swell. Then pure DNA is added into the competent cells in a water solution.
Transformation efficiency: How many transformants (recombinant plasmids) are created per a microlitre of foreign DNA introduced.
Translation: The process of translating an mRNA into a protein. Watch a clear animation of it by St. Olaf's college.
UTR: Untranslated region. UTR 5' and UTR 3' are found between a gene-coding region and its promoter / poly-A -tail.

No comments:
Post a Comment